Whole Genome Sequencing
Edited by Jessie, Jen Moreau, SarMal, Sharingknowledge
Whole genome sequencing is used to determine the entire DNA sequence of an organism. Whole genome sequencing reveals the organism's genotype. Knowledge of an entire genome sequence can help researchers determine the organization of genes, gene function, and the way genes work together. Sequencing the human genome has helped determine the molecular basis for many diseases. For example, whole genome sequencing can compare the genome sequence of someone with a disease to the genome sequencing of someone without, and identify the genes or nucleotides that are different in those with the disease versus those without the disease.
There is two genome sequencing methods:
Dideoxy (Sanger) Sequencing
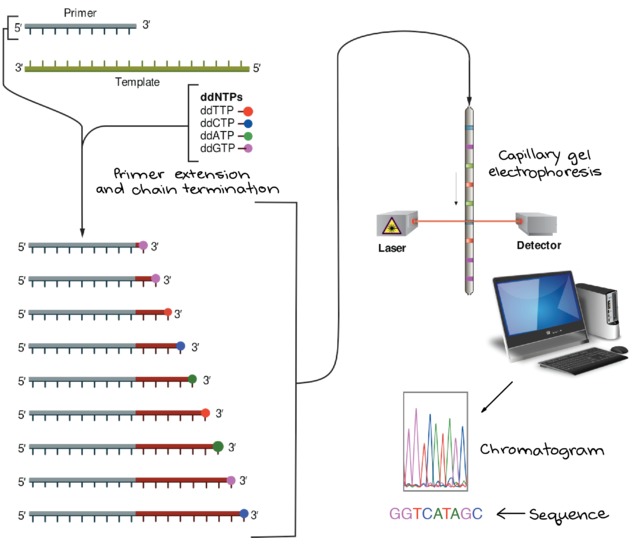
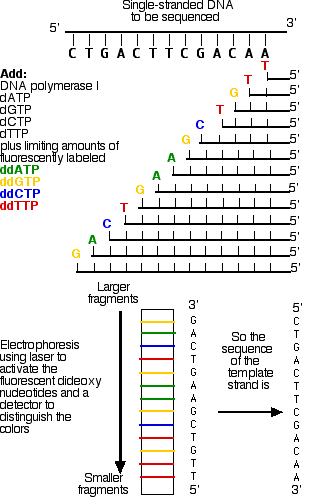
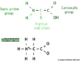
This method uses a denatured single strand of DNA, a DNA primer, a mixture of the 4 deoxyribonucleotide precursors (dNTPs), a mixture of 4 deoxyribonucleotide precursors (ddNTPs) that are labeled with different color fluorescent molecules, and a DNA polymerase. The N in NTPs refers to all of the nucleotide bases: adenine (dATP), thymine (dTTP), cytosine (dCTP), or guanine (dGTP), together they are referred to as dNTPs.
.
- 1Dideoxy (Sanger) Sequencing.Advertisement
- The polymerase, primers, dNTPs, and ddNTPs are mixed together.
- DNA polymerase synthesizes the new DNA in the 5' to 3' direction starting at the primer and stops every time the DNA incorporates one of the ddNTPs instead of the normal dNTPs. The ddNTPs have a single "H bound to the 3' carbon, instead of the usual "OH, so when a ddNTP is added, another nucleotide cannot bind and continue the chain and replication are stopped. Replication continues and produces a collection of DNA strands that were terminated randomly. This step produces a collection of DNA strands of varying sizes that all end with a fluorescently labeled ddNTP.
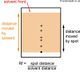
- The DNA fragments are then separated on an electrophoresis gel. An electrophoresis gel separates charged particles (like negatively charged DNA) by size. Shorter strands of DNA will migrate faster than larger pieces. A typical agarose gel isn't sufficient to separate something 1 or 2 nucleotides apart in size, so a polyacrylamide gel in a capillary tube is used. The fragments then move down the gel by precise size starting with 1 nucleotide, then two, then three etc. As the fragments move down the gel a laser shines light through the gel and excites the fluorescent molecules. A detector measures the fluorescence and displays which of the nucleotides was at the end of the strand. Reading the gel from the bottom up is the 5' to 3' orientation of the complement to genome sequenced (by base pairing rules, the original sequence can be determined). This method can produce a sequence of 500-750 nucleotides; it is generally used to sequence genes or a small genome.
Whole Genome Shotgun Sequencing
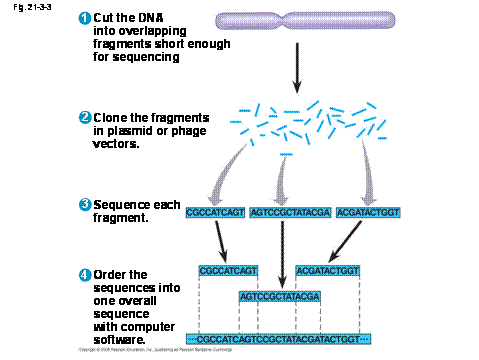
In this method, the sequence is first broken up and then sequenced.
- 1Whole Genome Shotgun Sequencing.
- Once the DNA has been purified, it is broken up into random sized overlapping fragments using restriction enzymes.
- Each fragment of DNA is cloned into a plasmid cloning vector, this step is called library prep. A library of all of the fragments in plasmids is made.
- The DNA from each plasmid is sequenced. This gives many short pieces of sequenced DNA, but now it needs to be assembled back to one long strand.
- The sequenced DNA fragments are put into a computer program that assembles them like puzzle pieces. The sequenced fragments of DNA are called reads. Overlapping reads are grouped together into contigs. Gaps between contigs are filled in by finding a fragment that overlaps both ends of nearby contigs. The ends of the pieces line up and are paired to create one contiguous piece, called a scaffold.
When working with DNA it isn't as obvious how the sequence lines up because there are only A, T, C, and Gs, but having many copies and various sized overlapping pieces makes it possible for a computer algorithm to put them back together.
More recent improvements in this type of sequencing and computer algorithms have made it possible to use this method to sequence the large genomes of eukaryotes. This is now the method of choice for sequencing all organisms.
Referencing this Article
If you need to reference this article in your work, you can copy-paste the following depending on your required format:
APA (American Psychological Association)
Whole Genome Sequencing. (2017). In ScienceAid. Retrieved Apr 29, 2024, from https://scienceaid.net/Whole_Genome_Sequencing
MLA (Modern Language Association) "Whole Genome Sequencing." ScienceAid, scienceaid.net/Whole_Genome_Sequencing Accessed 29 Apr 2024.
Chicago / Turabian ScienceAid.net. "Whole Genome Sequencing." Accessed Apr 29, 2024. https://scienceaid.net/Whole_Genome_Sequencing.
If you have problems with any of the steps in this article, please ask a question for more help, or post in the comments section below.
Comments
Article Info
Categories : Biochemistry
Recent edits by: SarMal, Jen Moreau, Jessie